Methods
Assay
Sample Collection
This project is conducted in the ICR mouse strain. Pregnant ICR mice for sample collection are purchased from Sankyo Labo Service Cooperation Inc. (Tokyo, Japan). At E9.5, 10.5 and 11.5, embryos are manually harvested into RNase-free cold phosphate-buffered saline (PBS) treated with DEPC (diethylpyrocarbonate) and fixed overnight in a solution of 4% paraformaldehyde in PBS at 4℃. Embryos were subsequently dehydrated through a graded series of methanol washes (25%, 50%, 75% and twice with 100%) in PBT (PBS containing 0.1% Tween-20) and stored at -20℃ until needed for WISH assays. If the expression of a given gene is found interesting, embryos at other stages were also collected for WISH assays.
Probe Synthesis
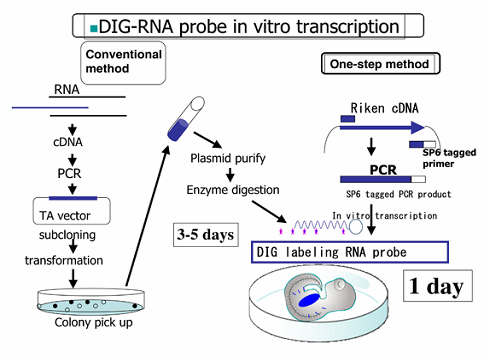
The conventional method of the probe synthesis includes bacteria culture and purification of DNA template, which are relatively lengthy for mass production of probes. To circumvent these obstacles, the one-step method for the probe synthesis has been developed. DNA fragments are amplified by PCR using plasmids as templates: primers are designed using Primer3 and PrimerExpress, and the Sp6 RNA polymerase promoter site is added to the 5' end of the reverse primer. Following the purification of amplified DNA fragments, probes are synthesized with Sp6 RNA polymerase according to the standard procedure in the presence of digoxigenin (DIG)-conjugated UTP.
Whole-Mount In Situ Hybridization
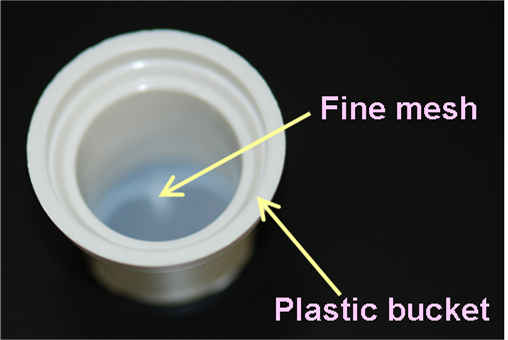


Whole mount in situ hybridization (WISH) assays are performed based on the standard protocol. All the procedure is manually handled, but 12-well plates and buckets with the fine mesh net (Netwell™ 12 well Carrier Kit for 15mm Inserts, Corning) are used, instead of vial tubes or centrifuge tubes, to expedite sample processing (see the figure). This enables each person to assay 24 genes (for 3 stages, which make a total of 72 embryos) at one time. The WISH assay performed at SMB includes the following steps:
- Bleach
- Protease K digestion
- Re-fixation
- Pre-hybridization
- Hybridization
- Post-hybridization washes
- Blocking
- Antibody reaction
- Antibody washes
- Color development
After satisfactory staining is developed, embryos are stored in 10% formalin in PBS at 4℃.
Microscopy
2D photo images
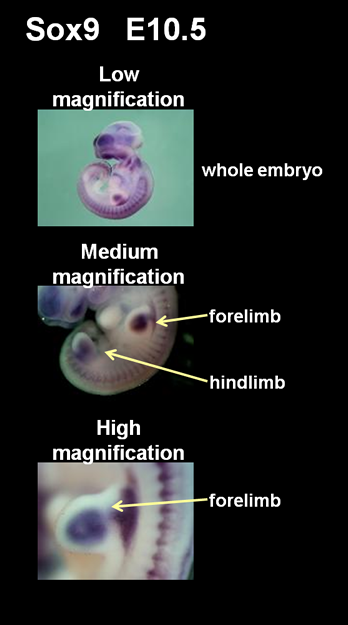
The colorimetric detection of hybrid signals are photographed under the light microscope (OLYMPUS SZX12) equipped with a CCD camera (OLYMPUS DP70). Specimens are fixed in agarose gel so that photos can be taken from any directions desired. When photographed, direct lighting is used and image processing (i.e. color balance, contrast) is controlled by the computer program (DP Controller, OLYMPUS). Generally, three photos are taken for each embryo: whole view of the embryo at a low magnification, the forelimb and the hindlimb at a medium magnification, the zoomed forelimb at a high magnification. Additional photos are taken if the gene expression is clearly detected in specific organs or tissues (i.e. brain, spinal cord, jaw, eye, nose, somite, heart, liver, tail) at appropriate magnifications.
AERO Images
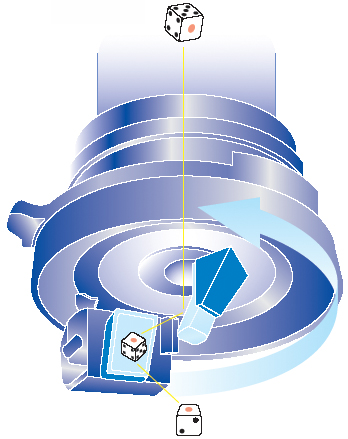
We have utilized an innovative microscopy System (HiRox, Tokyo) that enables us to capture 360-degree rotational images of the whole embryo. The key aspect of this system is the rotary head attached to the bottom of the body tube. This rotary head is equipped with two prism mirrors that allow the camera to capture the images tilted at 45 degrees to the microscopic stage. When an embryo is photographed, the rotary head laterally rotates by 360 degrees, capturing images at 2-degree intervals. This translates into 180 images photographed by a single shot, which together produce a motion picture by sequentially shown, termed AERO image. Researchers can easily view the rotational image, termed AERO image, at any lateral angle desired using a computer mouse by clicking and dragging around the edge of the screen and moving circularly.
The unique and critical features that make this novel recording system valuable include:

- Since the 360-degree view of the specimen is captured by the rotary head, significant amount of image data can be recorded by a single shot as opposed to the conventional 2D image, which is a mere snapshot of the one aspect selected by somebody else. This feature is highly favorable for the image data provided by the database as a whole embryo can be exposed to users in an unbiased manner. In particular, AERO images are useful to document gene expression in the regions that cannot be observed by a typical 2D lateral view, such as limb buds, genitalia and the maxillofacial region.
- AERO images can be easily obtained by a less laborious procedure, which is basically the same as that of conventional 2D images; an assayed embryo is placed under the microscope and serial images are captured automatically after manually focused. For this procedure, no particular treatment or processing is required unlike other 3D imaging systems. Thus, all the images are provided as primary data that are directly obtained from the embryo by a nondestructive method and images are highly reproducible regardless of individual expertis e. These features are advantageous to a large-scale assay in which numerous image data are produced.
- AERO images can be directly downloaded from the EMBRYS database to view without any special software. The image can be saved, copied, transferred and deleted like other commonly used formats in the user’s computer. This simple operation to handle AERO images will make it easy to use them as a research tool.
Annotation of Gene Expression Patterns
The expression of each gene is evaluated in order to determine where and how clearly it is expressed. Please refer to the embryo atlas below that demonstrates the anatomical structures focused for annotation.
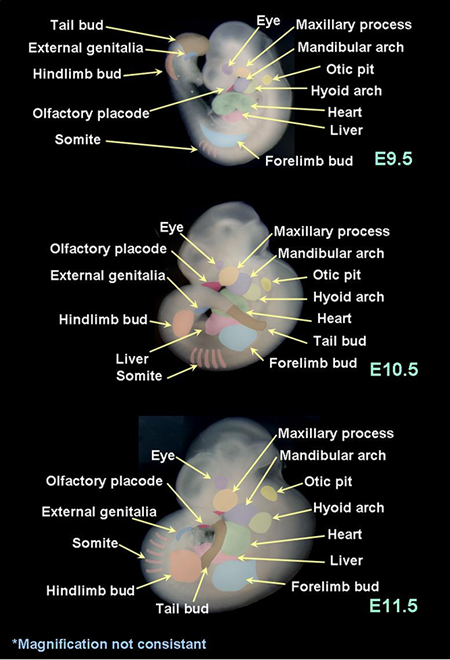
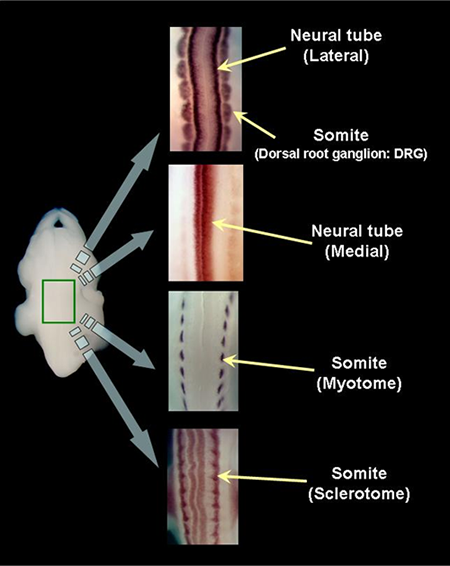
In the first column of the spreadsheet (1), whether any expression at any organs or tissues was detected or not is evaluated (y: yes, blank: no, or not determined). "y" in this column may include vague staining or ubiquitous staining resulted from our assay. In the next three columns (2), whether the relatively clear localization was detected or not is evaluated in each stage. If this column is marked "y", the organs or tissues where the expression was localized were further evaluated in the rest of the corresponding columns (3).
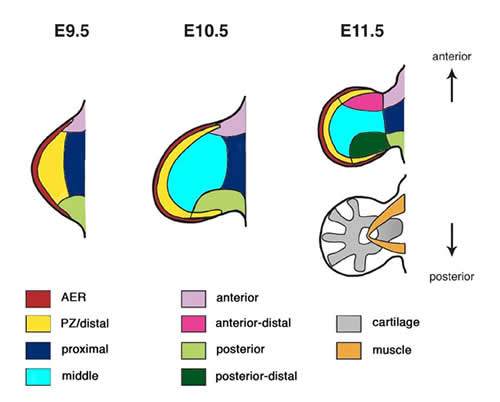
Since our original interest stems from limb development as an optimal model for the pattern formation, and the limb bud is readily observed under the microscope, the gene expression in the developing limb bud is particularly evaluated in detail (4).
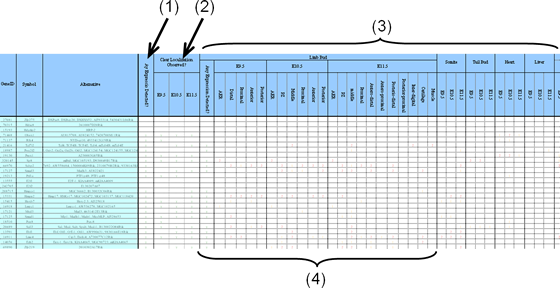
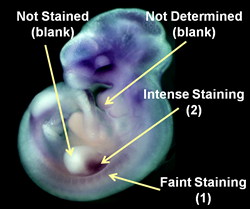
Although a WISH assay is not quantitative, each gene, in fact, manifests significant difference in the intensity of the colorimetric signals as a result of WISH assays. Thus, the signal intensity is also evaluated (2: intense signal, 1: faint signal, blank: no signal, or not determined due to the obscure staining).
Note that the annotation is provided as-is without warranty of any kind. Although the gene expression has been annotated by multiple experts, it is undeniable that such evaluation entails subjectivity to a certain extent.
