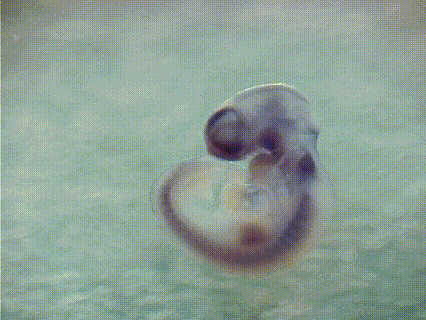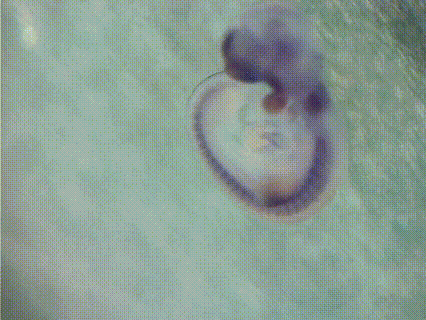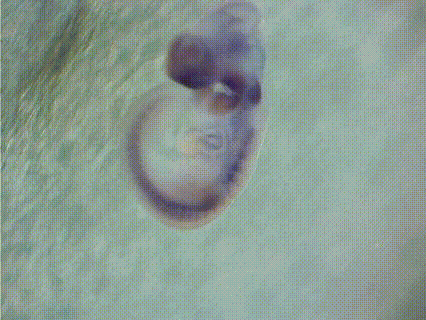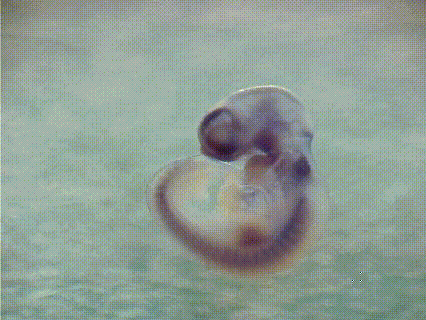What's new
Jun 11, 2025 : EMBRYS database is now hosted at Institute of Science Tokyo (Science Tokyo), established through the merger between Tokyo Medical and Dental University (TMDU) and Tokyo Institute of Technology.
Mar 25, 2017 : EMBRYS database was relocated to Department of Systems BioMedicine, Tokyo Medical and Dental University (TMDU).
Aug 30, 2013 : AERO system was published and open to the public. The AERO system: a 3D-like approach for recording gene expression patterns in the whole mouse embryo. PLoS One. 2013 Oct 16;8(10):e75754.
Jan 25, 2013 : EMBRYS was updated. Annotations of gene expression are now available.
Jan 25, 2013 : EMBRYS was linked to EMAGE.
Jun 8, 2010 : The research based on EMBRYS was published in PNAS. The Mohawk homeobox gene is a critical regulator of tendon differentiation. PNAS 107(23):10538-10542, June 8, 2010.
Dec 15, 2009 : EMBRYS 1.0 (primary version) is now available.
Dec 15, 2009 : EMBRYS was featured on the article published in Developmental Cell, December.
May 21, 2009 : EMBRYS β-version is now open to the public.
Sep 9, 2008 : Annotation of gene expression is now available.
Aug 29, 2008 : EMBRYS β-version is now open!

The details of EMBRYS database are described in the following article:
Please cite this article as a reference of EMBRYS database!
Welcome to EMBRYS!
EMBRYS is a digital atlas of in situ gene expression patterns in the developing mouse embryo constructed by the Systems BioMedicine Laboratory (Asahara's Lab) at the National Center for Child Health and Development and now maintained at Institute of Science Tokyo (Science Tokyo). This database contains the expression patterns of 1,520 transcription factors (TFs) and TF-related factors in mouse embryos at E9.5, 10.5, and 11.5. Expression patterns are determined by whole-mount in situ hybridization with digoxygenin-labeled riboprobes. [See more...]
To retrieve gene expression patterns, searches can be initiated by entering the information in the box located at the top of this page, and click "Search". Queries can be made using either gene symbol or NCBI Gene ID. Following query, the images are displayed by clicking the desired gene symbol listed in the box. [See more...]
Annotation of gene expression was evaluated on each tissues and organs (see Methods: Annotation of Gene Expression Patterns) and the data can be downroaded from here (MS-Excel file).